For a project, I need to store large amounts of documents and retrieve chunks based on their relevance. My current solution is a small embedded Chroma, and I wanted something more scalable. Enters: Elasticsearch.
The code
For reference: all the code is available on GitHub.
Design
I’ll start with an explanation of the design. I want to implement a multi-level repository where the engine searches among summaries and then goes into the details. It is not a new strategy. For example, this is a paper discussing exactly the same idea.
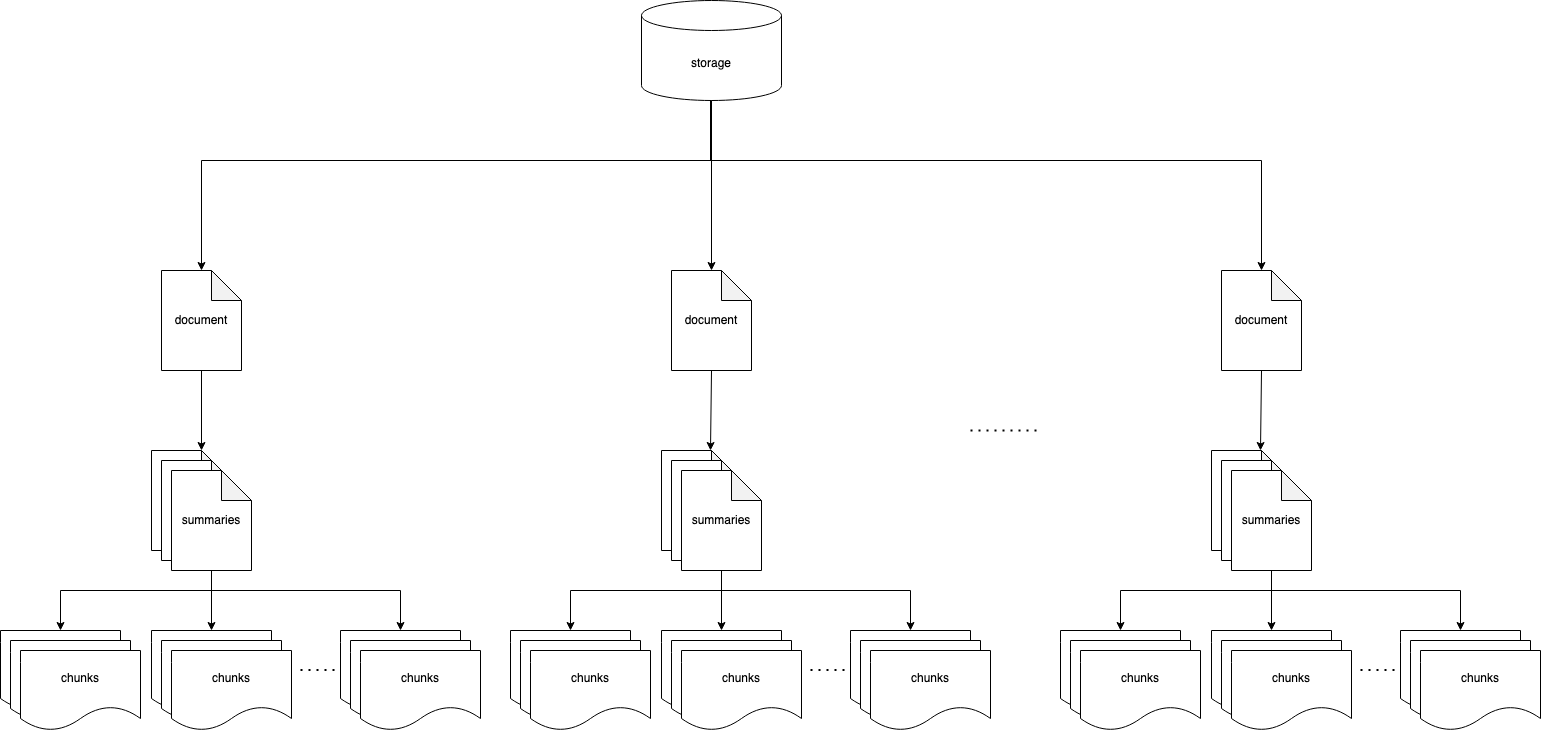
The diagram shows the logical model: documents are broken down into multiple summaries, and every summary is related to multiple chunks. Technically, summaries and chunks belong to different indexes.
When breaking down a long document in chunks, a small chunk may never mention the main topic and be found when researching that topic. With this method, chunks linked to a relevant block of chunks get a visibility boost.
It is worth to mention that there are alternative strategies, such as appending the higher level summary to the chunk or tagging.
Components
PDF reader with LLMSherpa
The first step is to break down a document into chunks. I will use a PDF parser based on NLM-Tika because it is layout-aware. Layout-aware parsers can recognise a column layout and create coherent chunks corresponding to an entire paragraph. They are not perfect, but they are much better than simple parsers that put all the text in a pipe and cut the chunks when they reach a certain length.
I used SimpleDirectoryReader from LlamaIndex as a base class. I override the standard PDF reader with one that I created. My reader implements a method load_data that calls LLMSherpa to do the heavy lifting.
def load_data(
self, file: Path, extra_info: Optional[Dict] = None
) -> List[Document]:
abs_path = file.absolute().as_posix()
doc = self.layout_pdf_reader.read_pdf(abs_path)
docs = []
last_header = None
for idx, chunk in enumerate(doc.chunks()):
parent_text = chunk.parent_text()
text = chunk.to_context_text(include_section_info=False)
keywords = self.keyword_model.extract_keywords(text,
keyphrase_ngram_range=self.keyphrase_ngram_range,
use_mmr=True,
diversity=self.keyphrase_diversity)
header = self.header_cleaner.cleanse(parent_text,
last_header) if self.with_header_cleansing else parent_text
last_header = header
metadata = {
'header': header,
'chunk_index': idx,
'file_name': file.name,
'keywords': keywords
}
docs.append(PDFDocument(text=text, metadata=metadata))
return docs
The result also contains headers that follow the structure of headers in the PDF. Unfortunately, the library makes lots of mistakes so, more as an experiment than anything else, I wrote some code to clean it up. I also extract keywords with KeyBERT. In retrospect, I did not need these two components, but they were already in my initial implementation, and I left them there for future experiments. For this exercise, the essential part is just the text extraction.
Summary with Ollama and Mistral
To summarise the content, I leveraged Ollama with Mistral. I use my usual code to build an engine for RAG, and I use a vector retriever that does not do anything. It is unlikely that summaries may need a proper retriever in the future, but, in case, it would be as simple as replacing vector_retriever_chunk
def _get_instance(self) -> RetrieverQueryEngine:
if self._engine is None:
llm = Ollama(model=llm_model)
self._llama_debug = LlamaDebugHandler(print_trace_on_end=True)
callback_manager = CallbackManager([self._llama_debug])
service_context = ServiceContext.from_defaults(llm=llm,
embed_model="local",
callback_manager=callback_manager)
vector_retriever_chunk = NullRetriever()
text_qa_template = ChatPromptTemplate([
ChatMessage(
role=MessageRole.SYSTEM,
content=system_prompt,
),
ChatMessage(
role=MessageRole.USER,
content=user_prompt,
),
])
self._engine = RetrieverQueryEngine.from_args(
vector_retriever_chunk,
service_context=service_context,
verbose=True,
response_mode=ResponseMode.COMPACT,
text_qa_template=text_qa_template
)
return self._engine
The rest of the logic is a simple call to the LLM with a prompt asking to summarise the content.
def summarise(self, text: str, debug: bool = False) -> str:
engine = self._get_instance()
response = engine.query(text)
if debug:
LLMSummary._print_debug(llama_debug=self._llama_debug, response=response)
return response.response
Storage with Elasticsearch
I want to use Elasticsearch as a repository for my embeddings, so the first thing is to create an index with a dense_vector that is essential for a k-nearest neighbor (kNN) search.
def create_index(self, index: str):
self._es().indices.create(index=index, mappings={
'properties': {
'embedding': {
'type': 'dense_vector',
}
}
})
I use SentenceTransformer to create embeddings. The method that adds the documents to the database makes it transparent.
def _get_embedding(self, text: str):
if self._model is None:
self._model = SentenceTransformer(sentence_transformers_model_name)
return self._model.encode(text)
def add_document(self, document: dict, index: str) -> str:
document = {
**document,
'embedding': self._get_embedding(document['text']),
}
response = self._es().index(index=index, body=document)
return response['_id']
The find method uses a mixed technique. The kNN query is used to find the candidates closest to the query, and there is an optional filter to limit the research among nodes linked to specific parents.
def find(self,
query: str,
index: str,
top: int = 10,
parent_ids: list[str] = None):
query_filter = {'filter': []}
if parent_ids is not None:
query_filter['filter'].append({
"terms": {
"parent_id.keyword": parent_ids
}
})
return self._es().search(index=index,
knn={
'field': 'embedding',
'query_vector': self._get_embedding(query),
'num_candidates': top, # shard level
'k': top, # total
**query_filter
})
Putting everything together
Ingestion
The ingestion is in three parts:
- The ingestion uses the reader to extract a list of lists of chunks (one list per document).
- The chunks are grouped in blocks, chained in a single text, and summarised by the LLM.
- The summaries are stored as individual documents in the index
summary, and their respective chunks are stored in the indexchunkswith aparent_idattribute.
def ingest(self, folder: str):
reader = PDFDirectoryReader(llm_sherpa_url=llmsherpa_api_url,
input_dir=folder,
recursive=True,
with_header_cleansing=True)
for document_chunk_list in reader.iter_data():
for chunks in self._chunks(document_chunk_list):
text = '\n'.join([d.text for d in chunks])
summary = self._llm_summary.summarise(text)
doc_id = self._elasticsearch.add_document(document={
"text": summary
}, index=self._index_name_summary)
for chunk in chunks:
self._elasticsearch.add_document(document={
"text": chunk.text,
"parent_id": doc_id,
}, index=self._index_name_chunks)
Search
The search reflects the ingestion:
- Research the index
summaryto find the top results. - Research the index
chunksto find the top results linked to the top summaries.
def find(self,
query: str,
top_summary: int = 3,
top_chunks: int = 10):
summary_results = self._elasticsearch.find(query=query,
index=self._index_name_summary,
top=top_summary)
ids = [r['_id'] for r in summary_results['hits']['hits']]
chunk_results = self._elasticsearch.find(query=query,
index=self._index_name_chunks,
top=top_chunks,
parent_ids=ids)
return [chunk['_source']['text'] for chunk in chunk_results['hits']['hits']]
Conclusions
To try it, store PDF documents in ./pdf, download the code from my repository, and start the external services with start-dependencies.sh (Docker and Ollama are required), and then run app.py.
The application will index the PDFs and run the query in app.py.
The output will look as follows:
parents:
- id:_AseKI4BGKouGWn47Kre
score:0.52408326
- id:SAsgKI4BGKouGWn4aat0
score:0.5158203
- id:8gseKI4BGKouGWn4pao4
score:0.5073944
...
chunks:
- id=MQsgKI4BGKouGWn4Pavt
parent_id=8gseKI4BGKouGWn4pao4
score=0.6156311
text=
"""
foo bar
"""
...
